IHC Stain Quantification
9/30/2017
My lab was doing an analysis of breast cancer patients looking for correlations between cytotoxic T cells and overall survival. Part of this involved investigation of IHC tumor punches and panel of antigens.
An area for improvement was in the “stain quantification.” We were judging the stain coverage of the stains by eye using a human-rating system, 1-5, to indicate “how stained” a slide was. Surpisingly, this is a common methodology (Taylor, 2006).
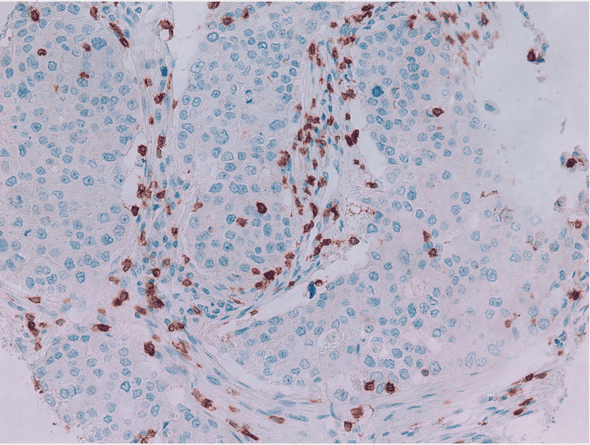
In research, I found some existing programmatic solutions to the problem (Varghese, 2014, Shi, 2016) but they were either not high-throughput, proprietary, or plugins for ImageJ (heavy). Given that I wanted to find “how-stained” each IHC slide was for >300 images, nothing looked ideal.
Solution
IHC Quantification script: github.com/JJTimmons/misc/tree/master/IHC_Quant
My naive solution was brightness thresholding using the python cv2 module. Strained regions should be darker than their unstained counterparts.
To find the brightness cutoff to use for a “stained” region, I uploaded a few sample images to ImageJ and played with the built-in thresholding tools. HSV format was particularly suited for this type of thresholding since saturation is built into the image format.
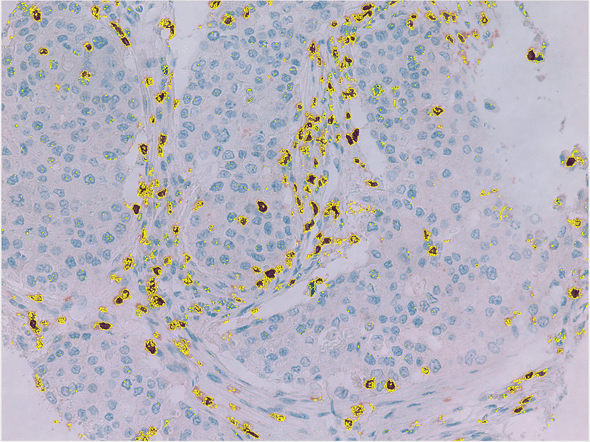
This was a good start, but the other factor to take into account was that not 100% of the slides were covered by tissue. Fortunately, since even the thin layers along the edges are slightly darker than the areas without any tissue in the slides, I was also able to threshold on tissue as well.
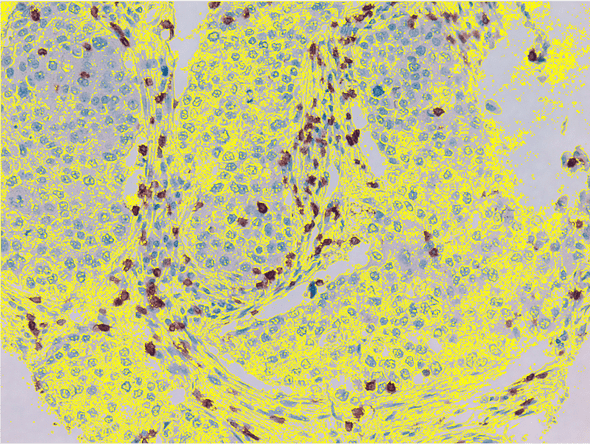
With these two counts in hand (the number of pixels that are “stained” versus the number that fall under the class of “tissue”), it was possible to create a ratio for each slide.
Code
First, I quantified the number of stained pixels in the saturation matrix of the HSV image.
import numpy as np
CELL_CUTOFF = 209
STAIN_CUTOFF = 147
def count(img, cutoff):
img = img[:,:,2]
sumd = len(img[img<cutoff])
return sumdLogical array indexing with numpy was a significantly fastest approach that looping through all the pixels. I then found the number of pixels above the two thresholds (one for tissue and one for stain).
import cv2
img = cv2.imread(d + "/" + im)
img = cv2.cvtColor(img, cv2.COLOR_BGR2HSV)
st = count(img, STAIN_CUTOFF)
ce = count(img, CELL_CUTOFF)
per = st / cecv2 has a built-in definition for converting an image to the HSV format that’s convenient for the cut-offs.
from __future__ import division
import os, sys
import cv2
import numpy as np
print "IHC start"
CELL_CUTOFF = 209
STAIN_CUTOFF = 147
def count(img, cutoff):
img = img[:,:,2]
sumd = len(img[img<cutoff])
return sumd
# loops thru each dir
fd = open("stainRatio.dat", 'w')
results = ""
ind = 0
di = os.walk('.').next()[1]
s = [st.split(" ")[1] for st in di]
for d in [y for (x, y) in sorted(zip(s,di))]:
results += "\\n" + d + "\\n"
# sort on slide number
files = os.walk("./" + d).next()[2]
end = [e.split(" ")[0] for e in files]
# loops thru each file
for im in [f for (e,f) in sorted(zip(end,files))]:
img = cv2.imread(d + "/" + im)
img = cv2.cvtColor(img, cv2.COLOR_BGR2HSV)
st = count(img, STAIN_CUTOFF)
ce = count(img, CELL_CUTOFF)
per = st / ce
results += im + " " + str(per) + "\\n"
ind += 1
print("done with " + str(ind) + " of " + str(len(di)))
fd.write(results)
fd.close()The full script (as configured above) will walk through a directory of patient records, quantify the IHC stain % of each image, and save it in a dat file (though lots of this is specific to the directory format being sent, for example: each patient directory is something like “Patient 1”, so I split+sort on folder name).
Caveats
This script would be better were the thresholds not hard-coded. A slightly brighter overall IHC slide might mis-classify tissue as empty fixative. A better solution would be to present the user with a sample image and asked them to pick a representative stained area and an unstained area, then find the average saturation in a blotch around both points (using that to calibrate the threshold for a given image set)
A much better approach would be to find a ratio of stained cells, rather than to use surface area as a proxy. The shape of a cell could be learned and the outlines could be drawn around each cell, after which saturation within the boundaries could be used to determine staining on a cell by cell basis.